This post is the first of six deep dives I plan to do into the significant technology challenges we have solved at Truveta using the power of AI to enable healthcare research using real-world data. Today I would like to talk about our approach to answering: How to generate and sustain data gravity in a consortium of healthcare data?
What does data gravity mean in Truveta’s context? If the contribution of healthcare data into our consortium by member healthcare systems gives increasing benefit to all users and members, encouraging them to bring in even more data, then the system has positive reinforcement, aka “data gravity.”
Truveta Embassies enable data gravity
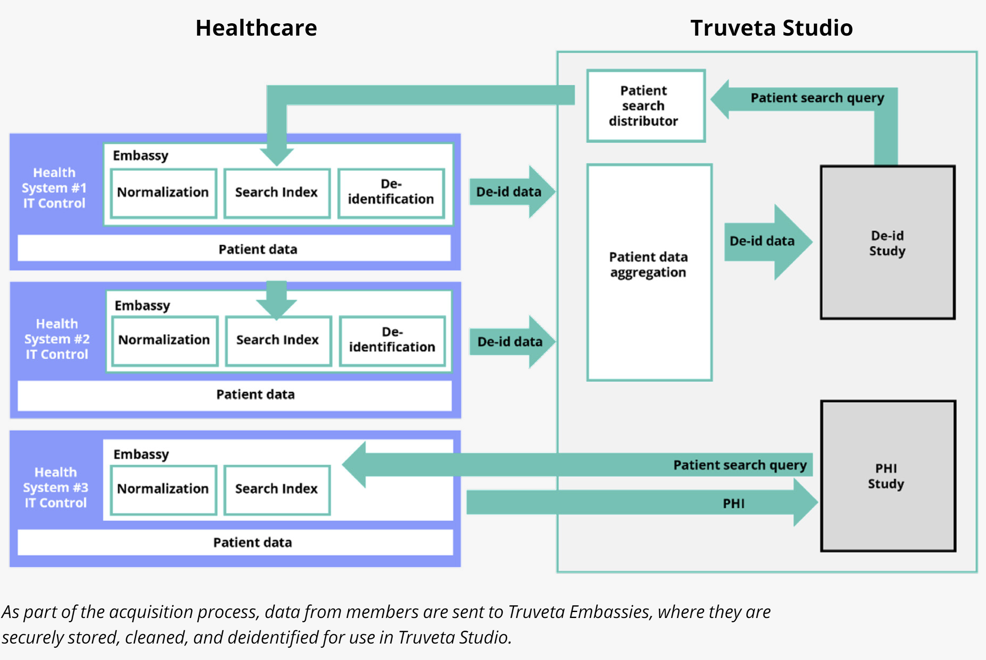
Truveta Embassies are secure environments in the cloud owned by the respective health system members where they can bring in their source healthcare data. This data includes Electronic Healthcare Records (EHR) of clinical events generated from line-of-business (LOB) apps, claims data, clinical notes, images, genomics, and more. (See the Truveta white paper “Our approach to data quality” for more details on Truveta Embassies.)
All this incoming data can be in a variety of formats and schemas, and is, at best, semi-structured. This is because members use a variety of applications and instrumentation in their healthcare operations, and the data is recorded by busy clinicians who may use multiple variations, abbreviations, and introduce ambiguities.
The challenge of data normalization
Firstly, to make this data available to researchers in a seamless manner such that they do not have to worry about the diversity and messiness at source, this data needs to be normalized to a standard schema called the Truveta Data Model (TDM) through data transformations. The basic information elements used in expressing our knowledge about patients, their conditions, treatments, medications, lab results, etc. are the standard clinical concepts defined in various medical ontologies such as SNOMED CT, RxNorm, and LOINC. These are important building blocks used by researchers to perform their research.
Secondly, we need to ensure that during these transformations, the data also accrues clinical accuracy as ambiguities are resolved. The quality and completeness of this data for each patient is critical. Only then can the data be indexed and made searchable, so that researchers can query for specific population cohorts of interest. Then they can access the data of the population in Truveta Studio after a deidentification process and use it as Real-World Evidence (RWE) of clinical care for clinical research.
Such research can lead to scientific breakthroughs and improvements in care delivery that benefit the entire ecosystem of life science and healthcare providers. Furthermore, the members can also use the same data for their own Health Economics and Outcomes Research (HEOR) and do comparative assessments and benchmarking of their operations against the broader industry standard. This produces a strong incentive for them to bring in more data.
So, it was clear to us that solving the data normalization challenge with accuracy and efficiency was a critical lynchpin for creating data gravity.
Could AI help?
The nature of the data sources, their diversity of concept expression, and bias towards billing rather than clinical accuracy made the problem challenging. We were advised by multiple experts that it’ll be difficult to use AI to transform the semi-structured data into their standard representation, as many such efforts have failed, and they suggested that we hire large teams of medical terminologists.
However, at Truveta, we persevered with the thesis that AI was the key to solving the challenge with excellence at scale and cost efficiency. Over three years, we purposefully invested in R&D to develop intellectual property and push the frontier. As a result, today we use state-of-the-art AI-based automation for a significant part of our data normalization pipeline. We started in early 2021 applying traditional Machine Learning (ML) to establish a baseline to compare improvements using more advanced technologies. We started with basic sub-word embeddings and continued improving from that baseline. The main improvements were seen after applying the full encoder-decoder text-to-text transformer (T5). We were already getting good results, which often surpassed the quality of human expert mappings for several clinical concept types. We compared our solution to the state-of-the-art (SOTA) for similar task which led to a publication in 2023 that was accepted at the 22nd International Semantic Web Conference ISWC-2023, Truveta Mapper: A Zero-shot Ontology Alignment Framework.
Eventually, we evolved to using large language models (LLMs) and large foundational models based on well-known decoder-only architectures, so that we have a generalist approach to solving multiple scenarios (aka domains such as diagnoses, medications, lab results, and specific groups of concepts therein). We will discuss the Truveta Language Model (TLM) and its uses in the next section. Similarly, for concept extraction from notes, we investigated a Transformer-encoder based approach in tandem with decoder/generative architecture, and later evolved it to accommodate large context sizes using a multi-stage concatenation approach.
Measuring the quality of AI
There were additional challenges with using AI. For example, industry experts warned us that even if we solved the challenge of scale through AI automation, the medical community may be reluctant to trust the results of AI systems since the technical metrics describing model quality may not be sufficiently informative to decide its fitness for clinical research. Therefore, we built our terminology team consisting of clinical experts from multiple areas of medicine that were instrumental not only in supervising our AI but also in the process of establishing expert baselines that would inform us about the level of decision quality that the medical community could trust. We have developed a Quality Reporting process for evidence generation and model certification that we can consistently apply to all models put into production and to assess model health on an ongoing basis. We will discuss more about this in our subsequent blog “Delivering accuracy and explainability.”
Compound AI systems – Agentic framework
One of the important discoveries we made along the way was the use of Compound AI systems to solve these complex problems. (Nowadays this is also referred to as an “agentic framework”.) The basic idea is to break down a complex task such as concept normalization from notes into several sub-tasks such as concept extraction, mapping, confidence assessment, active learning, and so forth, and then develop models for each of those subtasks that can work together as an orchestrated network to produce final answers. This approach has a much higher ROI than attempting to solve the entire task with one big model. It also allows us greater agility in addressing gaps as they are discovered. (Note that the components of the compound AI system are nevertheless generalist models that can work across multiple scenarios.)
Now let us look at those use cases in more detail. Specifically, we use AI in the following types of data transformations:
-
Semantic normalization (aka concept mapping) of semi-structured data
-
Concept extraction from clinical notes
-
PHI/PII detection and redaction from notes and images
-
De-identified patient matching
Semantic normalization of semi-structured EHR data
We use AI extensively in semantic transformations via the Truveta Language Model (TLM), a large-language, multi-modal AI model used to clean billions of daily Electronic Health Record (EHR) data points for health research. Using the unprecedented data available from members and the capabilities of Truveta’s clinical expert annotation team to label thousands of raw clinical terms, including misspellings and abbreviations, TLM is trained to normalize healthcare data for clinical research. TLM normalizes EHR data to maximize clinical accuracy and is trained without commercial bias, helping ensure research is conducted with data focused on clinical outcomes rather than billing.
Data on clinical events such as diagnoses of conditions, lab results, procedures, and prescriptions of medications are semi-structured. Clinicians use different terms based on their locale, training, and expertise. For example:
-
“Acute COVID-19,” “COVID,” “COVID-19,” “COVID infection,” and “COVID19_acute infection” (and hundreds of other variations) all refer to COVID-19
-
“600mg Ibuprofen” and “Ibuprofen 600mg” are the same thing
-
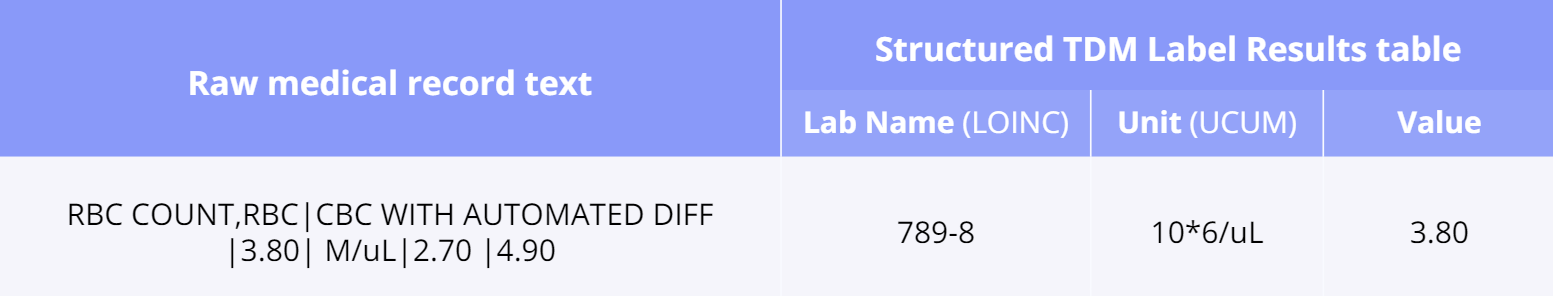
Even lab results may contain raw free text such as “RBC COUNT,RBC|CBC WITH AUTOMATED DIFF |3.80| M/uL|2.70 |4.90”
In other cases, clinicians may enter ontology-coded concepts but may map the same concept to different ontologies depending on their preference (for example, RxNorm or NDC for medication prescriptions). Thus, we need a mechanism for consistently mapping the unstructured strings to concept codes, as well as for aligning concept codes across ontologies. The aim is to ultimately map these various strings to the most correct and clinically meaningful concepts coded according to the standard ontologies, such as SNOMED, LOINC, UCUM, RxNorm, GUDID, CVX, and HGNC.
This is achieved by a specialized application of the Truveta Language Model (TLM) for concept mapping as well as ontology-modeling/alignment (see our paper Truveta Mapper). TLM treats cleaning the data as a translation task. Ontologies are represented as graphs, and the translation is performed from a node in the source ontology graph to a path in the target ontology graph. TLM leverages a multi-task sequence-to-sequence transformer model to perform alignment across multiple ontologies in a zero-shot, unified, and end-to-end manner. Multi-tasking enables the model to implicitly learn the relationship between different ontologies via transfer-learning without requiring any explicit cross-ontology manually labeled data. This also enables the formulated framework to outperform existing solutions for both runtime latency and alignment quality. Here is an example of a properly mapped and aligned raw string of lab results:
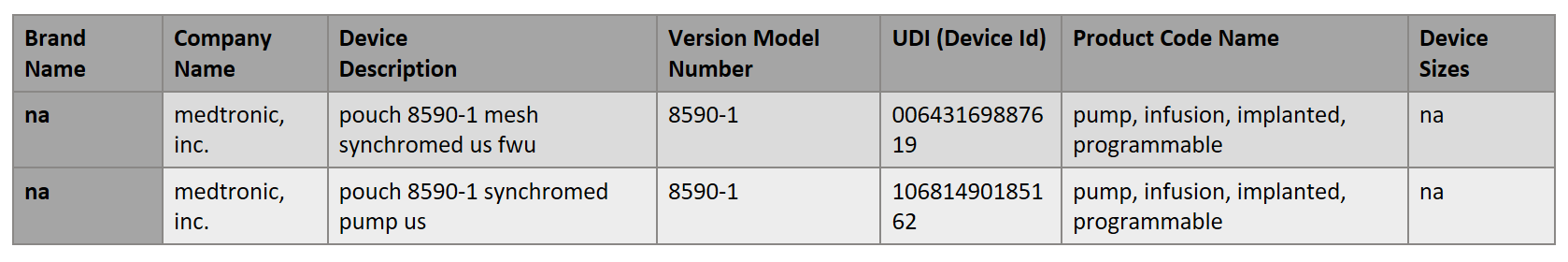
Another interesting and challenging area is the normalization of data about medical devices. The aim is to map each of the device health system terms to a Unique Device Identifier (UDI). The UDI code can be found in the Global Unique Device Identification Database (GUDID) which is administered by the FDA and is accessible here. Here is an example of the health system term in free text: “POUCH MESH DACRON – LOG63746 | MEDTRONIC – MEDT | 8590-1|”. The properly mapped version that TLM created would be (there are two possibilities):
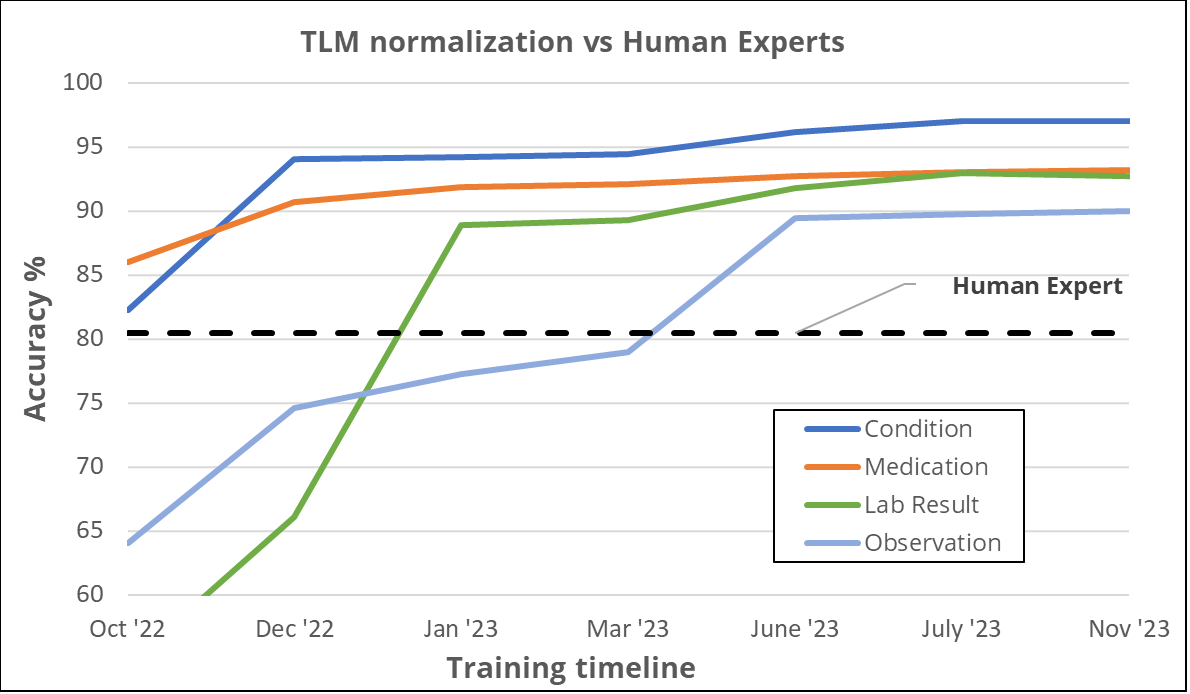
In general, our experience has been that after consistently tuning model performance through feedback from experts such as terminologists and clinical informatics, we are able to drive the performance of AI-based term normalization to levels that are at or above expert human performance. This is true across domains such as diagnosis, medication, etc. In some domains, such as devices, we are still in the process of achieving that goal but have made good progress.
Clinical notes and concept extraction
Clinical notes present an even more nuanced challenge. Since they are fully unstructured, we need to perform an additional task of concept extraction, where we first need to identify the parts of the text of the notes that refer to a particular concept. Then it can be followed by the mapping (normalization) of those spans of strings to concept codes, in the same way as we do for semi-structured elements. However, that involves further subtleties. One comes from the fact that the same span of text may refer to multiple concepts, and the spans may not even be contiguous. The other comes from the fact that researchers may want to extract custom concepts from notes that are relevant to their research but are not already defined in standard ontologies. So now we are not mapping, but really minting brand new concepts in our own ontology!
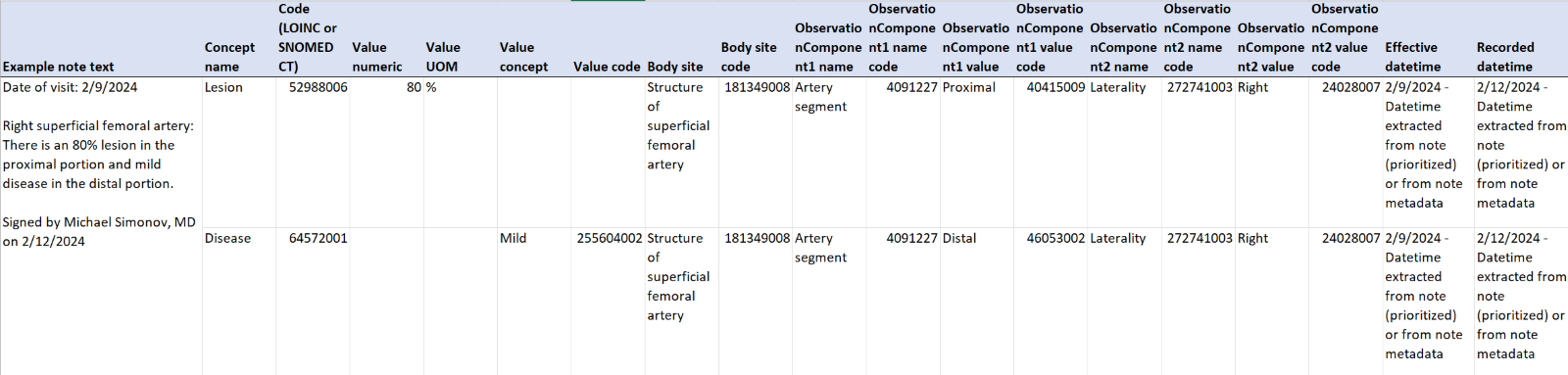
For example, the table below shows some clinical notes for patients with vessel disease. A note can result in multiple concepts such as “lesion” and “disease,” with body site locations and several observations in addition to date-time stamps.
To address all these requirements, TLM has been specialized to apply for concept extraction and mapping from notes. TLM identifies and normalizes clinical concepts within clinician notes, while accounting for typos/misspellings (e.g., “glipizisde” vs. “glipizide”) and clinical nuances such as negation (e.g., “patient denies feeling fatigued”), hypotheticals/conditionals (e.g., “Will consider starting low-dose glypizide if A1C still grossly elevated”), and family history (e.g., “Family Hx: Mother: Diabetes, Father/son: bipolar disorder”). TLM reasons over the entire medical record, accounting for changes over time to ensure the most accurate and complete information is structured. While we endeavor to use the generalist model where possible, in some cases we train (fine-tune) a scenario-specific model to achieve sufficient accuracy. Even in that case, the generalist TLM is used as a pre-annotator that helps human experts rapidly create the quantum of scenario-specific annotations needed for fine-tuning. (We will discuss domain adaptation, fine-tuning, and inference at scale for billions of notes in another blog post in this series, “Scaling AI models for growth”).
To support these various configurations of model ensembles, we are also actively investing in a framework for model versioning so that we can track changes and assure quality in the same way we do for our more general Software Development Life Cycle (SDLC).
PHI detection and redaction
An important consideration in enabling the use of AI models for concept pre-annotation, mapping, and extraction is the ability to train the models with deidentified (de-id) data. From a HIPAA point of view, if a model is trained on data with Personal Health Information (PHI), then the model and its outputs are also considered as having PHI. Working with such PHI models would severely restrict the agility of the model-building process, which requires significant iteration during training time.
So, the solution we devised was “train with redacted data, while inferencing on PHI data.” That is, for training the models we first redact all personal identifiers from the sample of the data (such as clinical notes) taken from embassies. Then that redacted data is brought into a separate zone and used to develop the “de-id models” (i.e., extraction and normalization models trained on de-id data). We also use techniques like surrogation and perturbation to make the data resemble synthetic data without losing the information required for model training. Once de-id models have been built, tested, and certified, they can be used to apply for inference on the PHI data in the respective embassies as part of the data transformation pipeline. Since the concepts extracted from the PHI data are mapped to ontologies, they are non-PHI. Furthermore, every data snapshot obtained from the Embassies and brought to Truveta Studio for clinical research undergoes a separate de-id process (which we will address in the blog post Ensuring privacy and compliance).
You may ask: How do we do the PHI-redaction of data that is then used for training AI models? Of course, through automation using another AI model! This PHI-redaction model is a variant of the concept extraction model for notes, with the target concepts being personally identifiable information rather than clinical concepts. The training of this PHI-redaction model is done in a highly secure and controlled environment (distinct from what is used for de-id model training), where a sample of PHI data is brought in a controlled and auditable manner, and then annotated by Risk Analytics experts, and that supervision is used to train and evaluate the redaction model.
One final point I would like to mention is our use of AI models for another crucial task in the data transformation pipeline called “patient matching.” If patients are double counted, it can introduce biases in population-level measurement of rates of outcomes such as adverse events because they skew the population size (denominator) as well as the outcome’s prevalence (numerator). Therefore, we do probabilistic patient matching over the de-identified data. We achieve this through an AI model that produces a “similarity” score for patient records based on secure hashes generated from their demographic and identifiable data. These hashes and scores are themselves fully de-identified. Using this, we cluster the deidentified records that have a similarity score above a certain threshold.
In other words, we are grouping or clustering the records and associating each cluster with a patient. This process is tuned for high precision. That is, when two patient records are associated in a cluster, the probability that they are the same person is very high (essentially close to 1.0). While assuring this precision, the recall of the algorithm (its probability to successfully join two records of the same person) is still quite high, so it makes a real impact on the quality of the research.
Conclusion
AI-based automation is used extensively at Truveta in critical functions of the data processing and transformation pipeline, leading to properly normalized patient records. As we saw in the introduction, doing this with clinical accuracy, efficiency, and scale was a prerequisite to achieving our goal of creating data gravity and, hence, was a key to the success of our business.
In the next post, I will talk about Delivering accuracy and explainability, where we address: How to ensure that the AI technologies used in our platform have requisite accuracy and explainability while assuring fairness and avoidance of bias.
Missed the first post in this series? Read it here.
Join the conversation on LinkedIn.