In this blog I will discuss our approach to scaling our AI technologies across multiple dimensions while controlling development costs, operational costs, and assuring stability. LLMs are a powerful new technology, but they also have large development, operational, and environmental costs. Hence, we need to use them in a judicious manner.
At Truveta, we continually make strategic decisions about modeling technology, size, training data, and knowledge retention policy (what needs to be remembered and what can be forgotten during pretraining for a specific task).
LLM development framework
As I discussed in my last post, we rely heavily on the “model graph” (aka “agentic framework”) approach that allows us to solve complex problems using a network of specially tuned small models (“agents”). By appropriately chaining and orchestrating such agents for solving a complex task, we can achieve high-quality results without having to train large models for every complex task. Furthermore, in addition to fine tuning, where possible we use prompt engineering as well as Retrieval Augmented Generation (RAG), which typically have a high ROI.
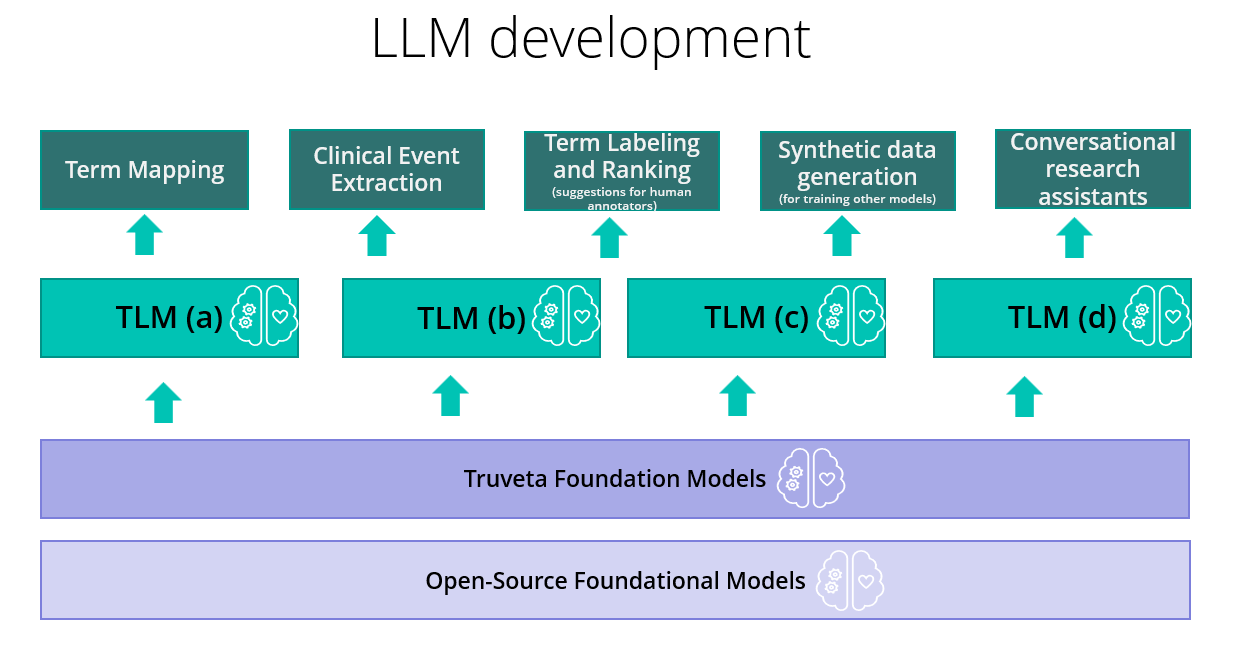
Our modelling stack is summarized in the above figure. We start with open-source foundational models and do domain adaptive pretraining with our proprietary healthcare data. Initially we started with text-based models, and we are now expanding multi-modal foundation models. We use these foundational models to fine-tune and align several different families of the Truveta Language Model (TLM). Each family is oriented towards solving a specific category of problems such as concept pre-labeling to aid human annotators, concept extraction, concept ranking to aid human annotators, concept normalization, synthetic data generation, and conversational research assistants. This whole stack needs to be optimized for scale. But scale is not one-dimensional; it has several distinct aspects, as we will now discuss.
Types of scale requirement and the tradeoff we need to make
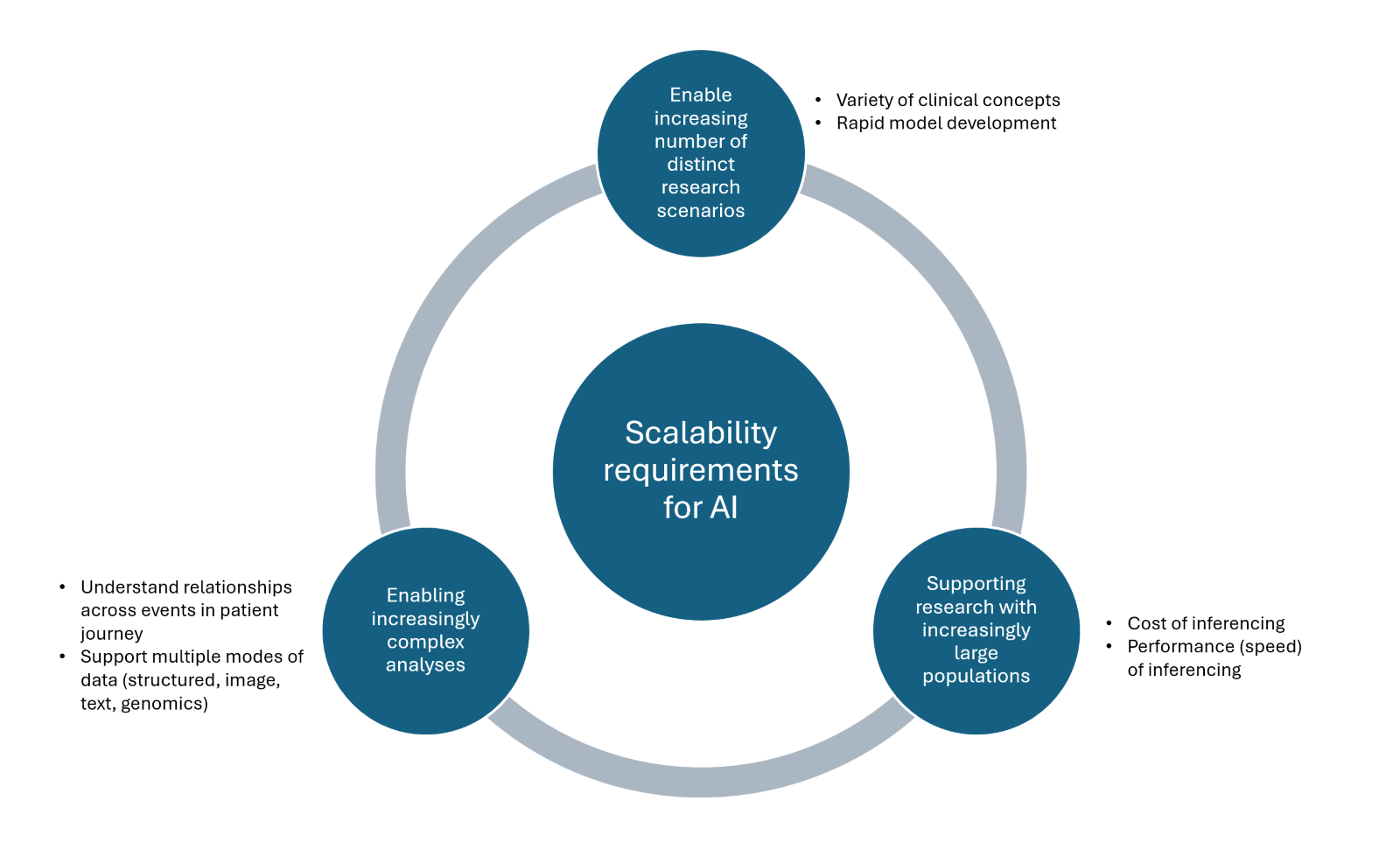
As shown in the figure above, to serve our customers’ needs, we need to be able to scale along three different dimensions:
- Scaling to support multiple types of research scenarios with distinct data needs
- Scaling to support research over large populations of patients
- Scaling to support increasingly complex analyses that use holistic aspects of a patient’s clinical journey
Scaling to many research scenarios
Any clinical research depends on certain core “must have” elements of normalized healthcare data such as conditions (diagnoses), lab results, medication requests and prescriptions, and observations.
In addition to these core needs, different groups of researchers may need normalized data pertaining to specific aspects of patient care and health operations. These may exist in ancillary EHR tables such as device use, or they may be derived concepts such as “mother-child relationships,” or they may even be custom concepts extracted from unstructured data such as “migraine frequency.” Even in a focused area such as cancer research, the specific data needs for various cancer types (and there are hundreds of them) vary significantly. The number of such additional research-specific data needs is continuously growing, and there may be relatively little reuse of those additional data across other scenarios.
This leads to a unique challenge: We want to scale AI models to support many increasingly distinct scenarios while making sub-linear investments in terms of model development effort.
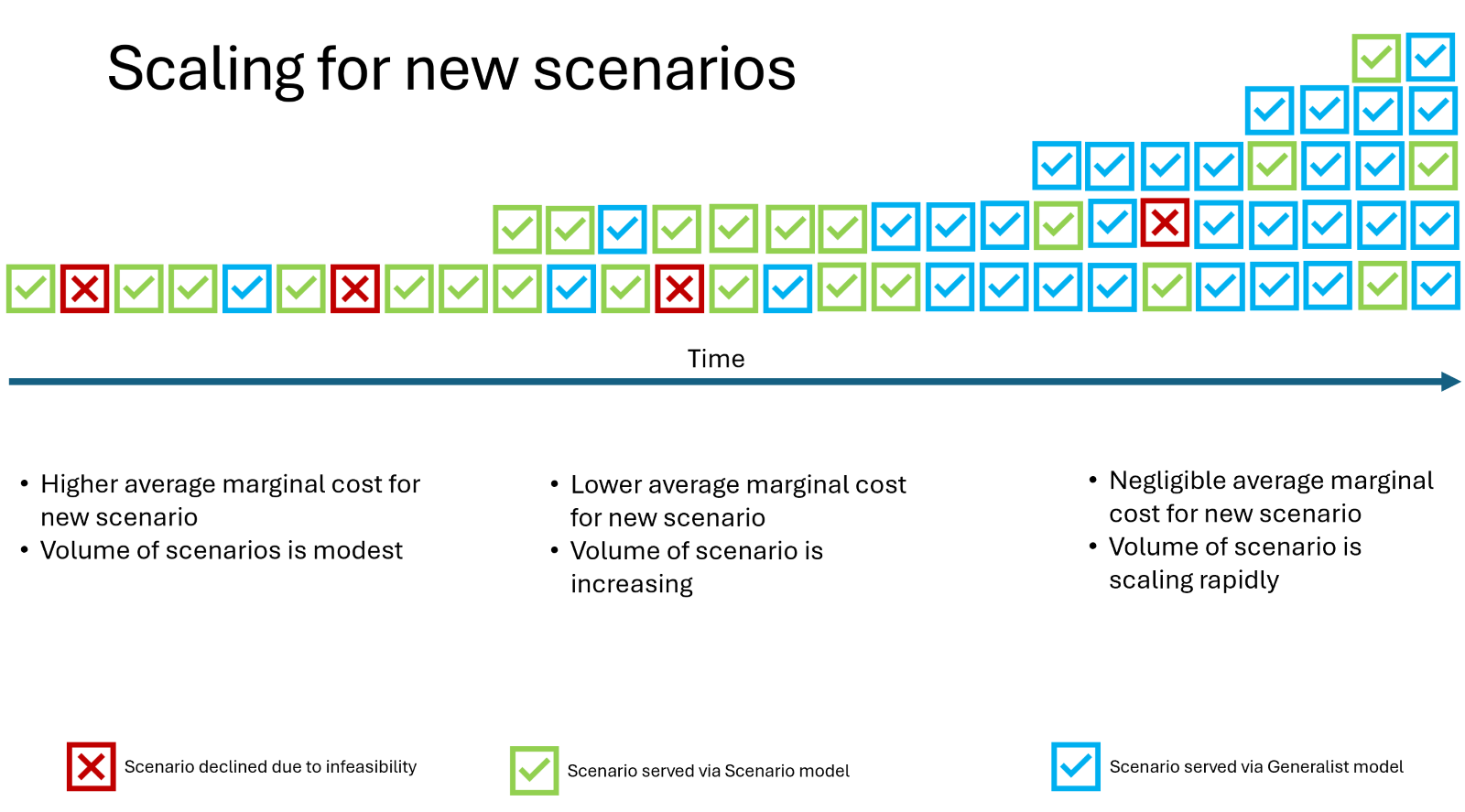
The primary approach here is to use generalist generative (decoder) models that need minimal extra supervision and training for new scenarios and can work primarily off “guidelines” provided by researchers themselves. In the case of prostate cancer, researchers can provide very specific guidelines for extracting and normalizing concepts from semi-structured and unstructured data pertaining to disease stage, site of metastases, histological grade group (Gleason score), PSA values, genetic markers and ECOG, and treatment history.
These cancer-type-specific needs may differ from cancer to cancer, but the pattern of providing guidelines that help tune the LLM for that specific task remains the same, and that is the key to scalability. The guidelines provide valuable prompting information, as well as helping generate high quality annotations (supervision) by human experts. These are used for domain alignment of our foundation models by prompt engineering and fine-tuning.
Scaling to large populations
The flip side of the previous requirement comes from the customers’ need to do research studies over large populations. Not all studies require data of large populations, but some do – for example studying effectiveness of widely administered COVID-19 vaccines or understanding clinical benefits of widely used GLP-1 receptor agonist medications easily runs into using data of tens of millions of patients. In these cases, using the generalist generative (decoder) model approach may simply be too expensive and time consuming at inferencing time because of the size and complexity of these models. Furthermore, due to dependence on expensive and scarce GPU resources, it also creates a vulnerability for the operation of the business.
In these cases, it makes sense to use scenario-specialized non-generative (encoder-only) LLMs that are cheaper and faster to execute (e.g. BERT-based). Of course, these models need more scenario specific training (which is not transferrable), and that results in a significant sunk cost per scenario. Another potential drawback is the small context length of such models, which would make them inapplicable to even modestly long clinical notes. However, we have done patent-pending innovation in extending the context length of such models by concatenation with a recurrent layer that allows fusion of the knowledge across long contexts, and essentially eliminates this drawback.
Inferencing with such models is very cheap and fast, and can be done without breaking the bank. We can easily scale to extracting and normalizing concepts from tens of millions of patients.
To reduce costs further, we also do several other optimizations in MLOPs, including dynamically using the smallest GPUs where possible, parallelization of inferencing workflows across VMs, and use of efficient inferencing frameworks.
Scaling for complex research analyses
Finally, the third dimension of scaling pertains to the researchers’ need to look holistically at aspects of the patients’ clinical journey spread across time and across multiple modes such as EHR, notes, images, genomics, proteomics, and more. Certain complex research analyses need to see the whole patient journey and understand the connection between events that may be widely separated in time: for example, exposure of a mother to certain medications during pregnancy and its result on her health in later life as well as the health of children in their adulthood.
Connecting events and concepts separated by long time intervals across different modes and people is very challenging. To achieve such knowledge extraction, we need to process very large context lengths (e.g. thousands of notes, large row-sets in structured data, large sets of images) and try to generate an entity-relationship graph of concepts across multiple events. This is still an open research area, and we are investigating modeling approaches involving patient journey summarization. The primary idea is to treat the patient journey of clinical events as a “document” in a special type of “EHR language,” and pre-train LLMs to learn the large-scale probability model of these documents by looking at data of millions of patients. To understand events pertaining to multiple modes of data (such as radiological images, EKR timeseries, and genetic testing), these models also need to have multi-modal capabilities. Once trained, such models can process patient journeys given in the “EHR language” and produce outputs in “human language” or in structured form like Truveta Data Model (TDM) that is suitable for analyses.
Conclusion
In summary, our use of AI models in EHR data harmonization and extraction of insights faces different types of scale challenges. While these requirements are typically at odds with each other, it is rarely the cases that they all need to be met simultaneously.
So rather than building a one-size-fits-all solution (which would be too costly and complex), we make strategic choices about the modeling technology, data, training/supervision effort, and knowledge retention policies to develop solutions that serve each challenge in an optimal way.
